The pmemd.cuda GPU Implementation
Por um escritor misterioso
Last updated 21 setembro 2024

Toward Fast and Accurate Binding Affinity Prediction with pmemdGTI: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration.

Fast Implementation of the Nudged Elastic Band Method in AMBER
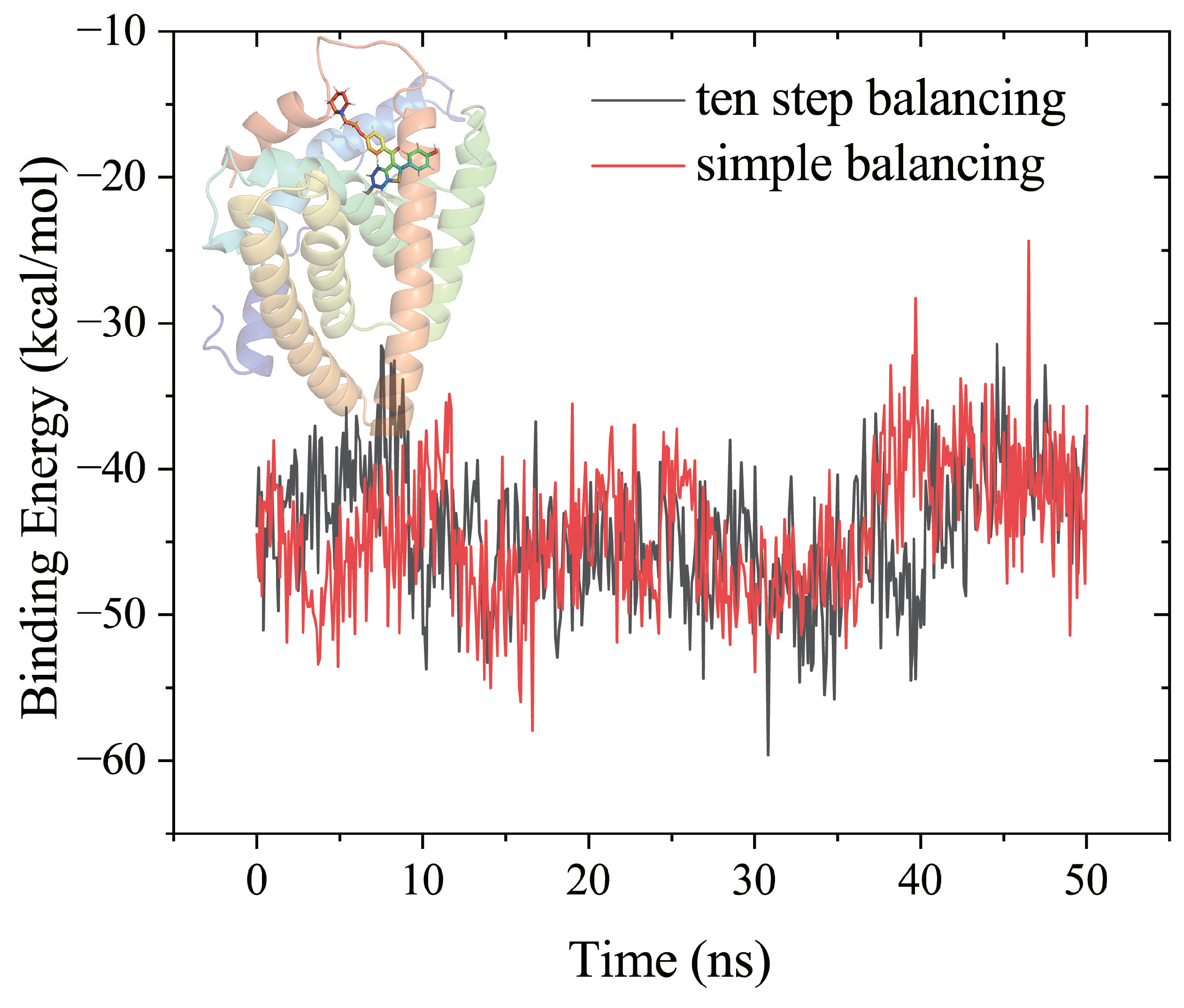
Biomolecules, Free Full-Text
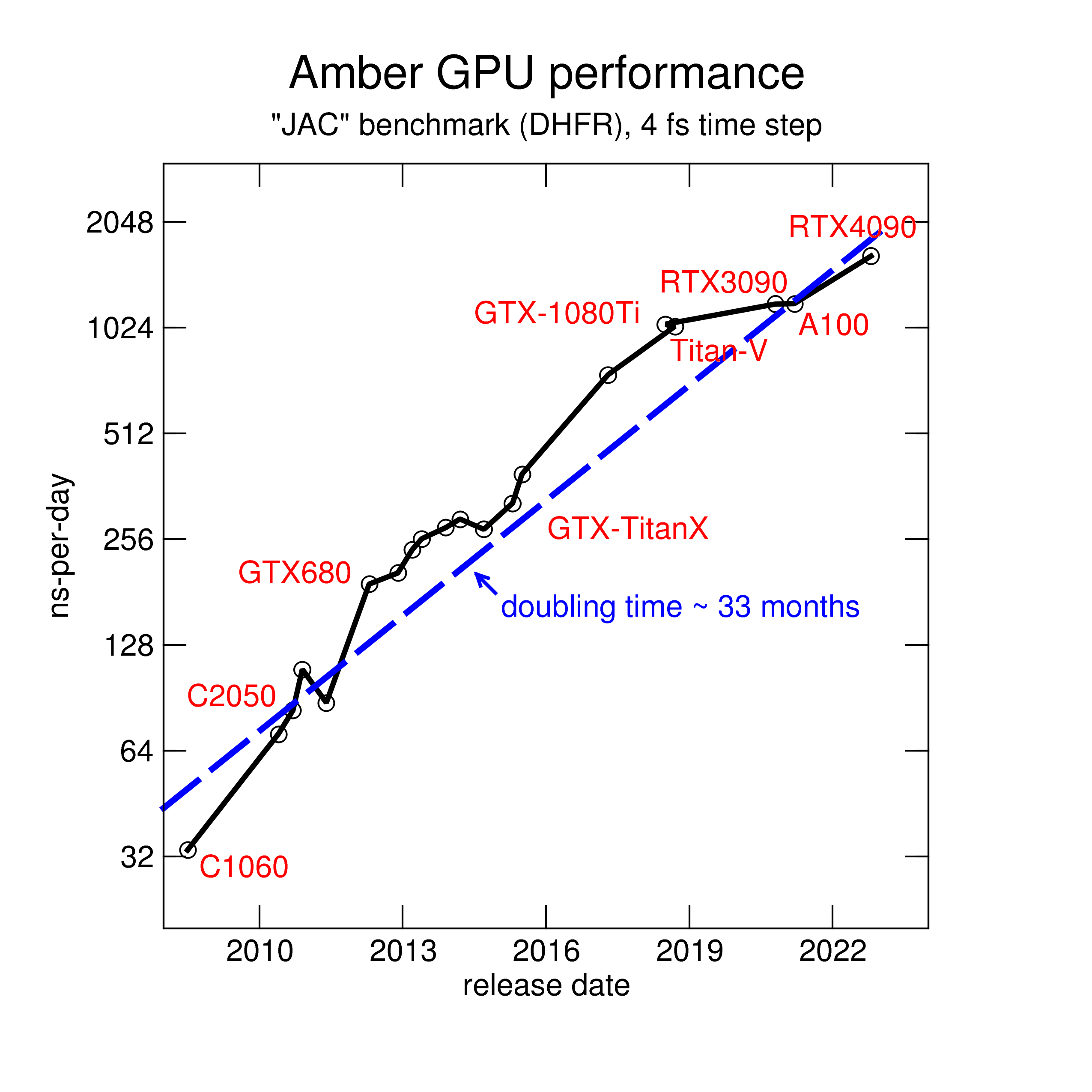
The pmemd.cuda GPU Implementation
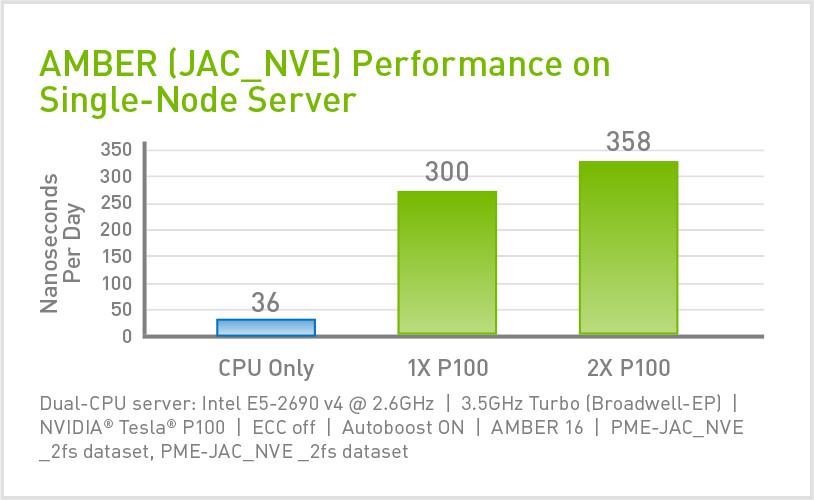
Amber Hardware & Software Configurations
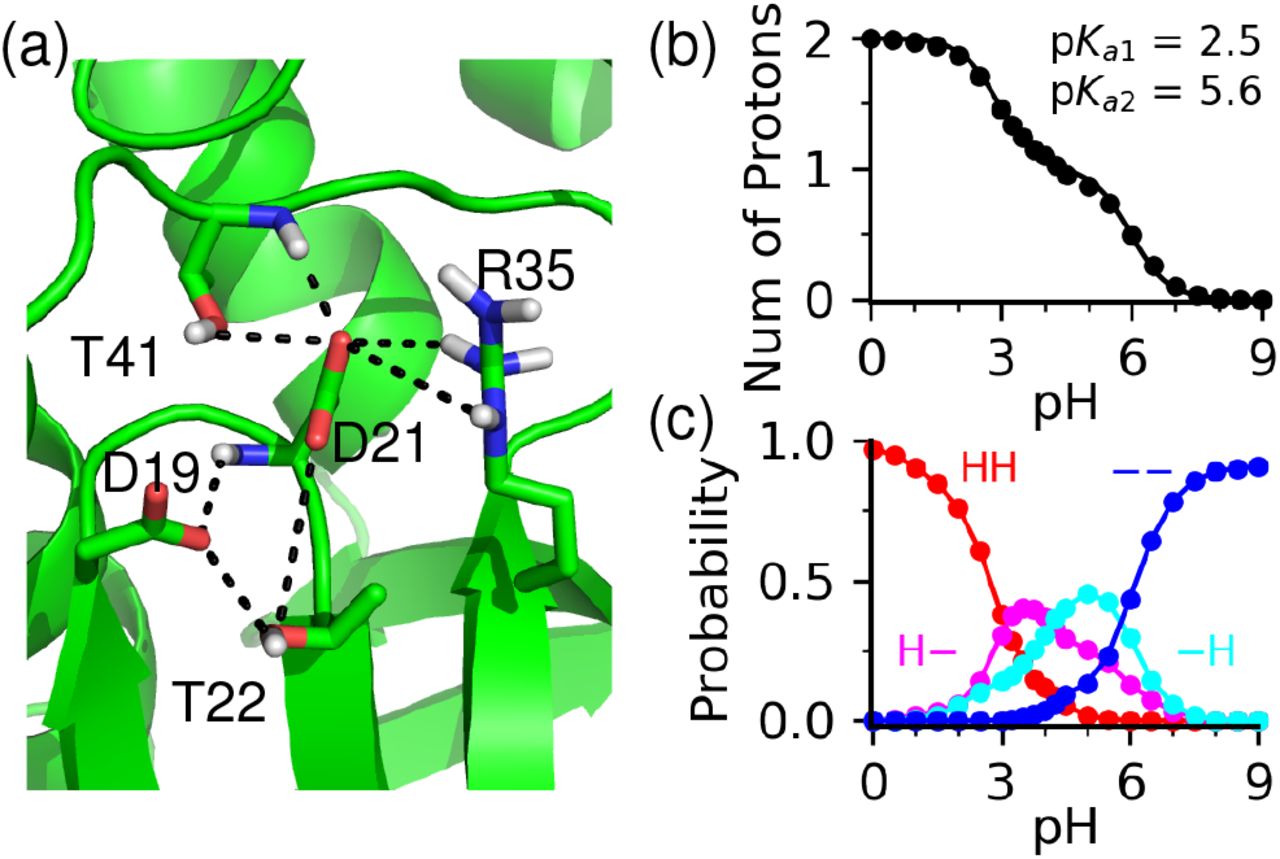
GPU-Accelerated All-atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber

Amber (PMEMD) GPU Support

Running AMBER on a GPU Cluster - Microway

GPU-Accelerated All-Atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber

Full article: Solvated and generalised Born calculations differences using GPU CUDA and multi-CPU simulations of an antifreeze protein with AMBER

Fast Implementation of the Nudged Elastic Band Method in AMBER

GPU-Accelerated All-atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber
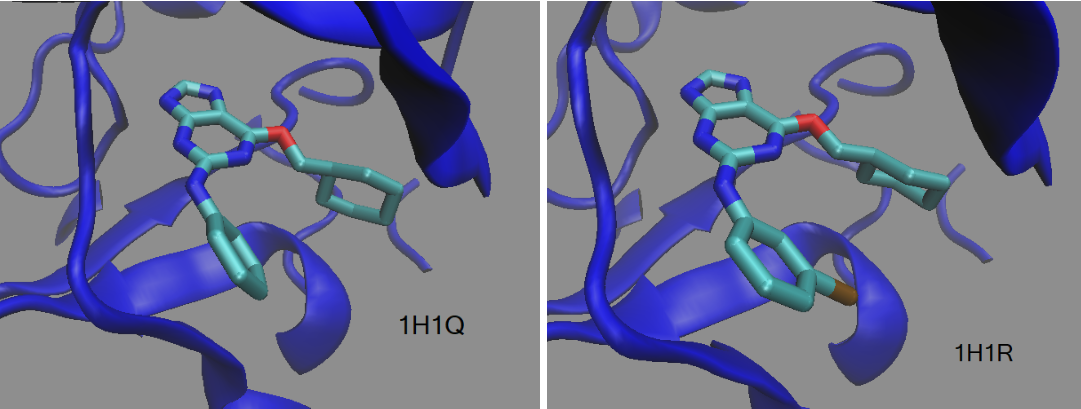
TI Calculation with DDBoost

Fast and flexible gpu accelerated binding free energy calculations within the amber molecular dynamics package - Mermelstein - 2018 - Journal of Computational Chemistry - Wiley Online Library
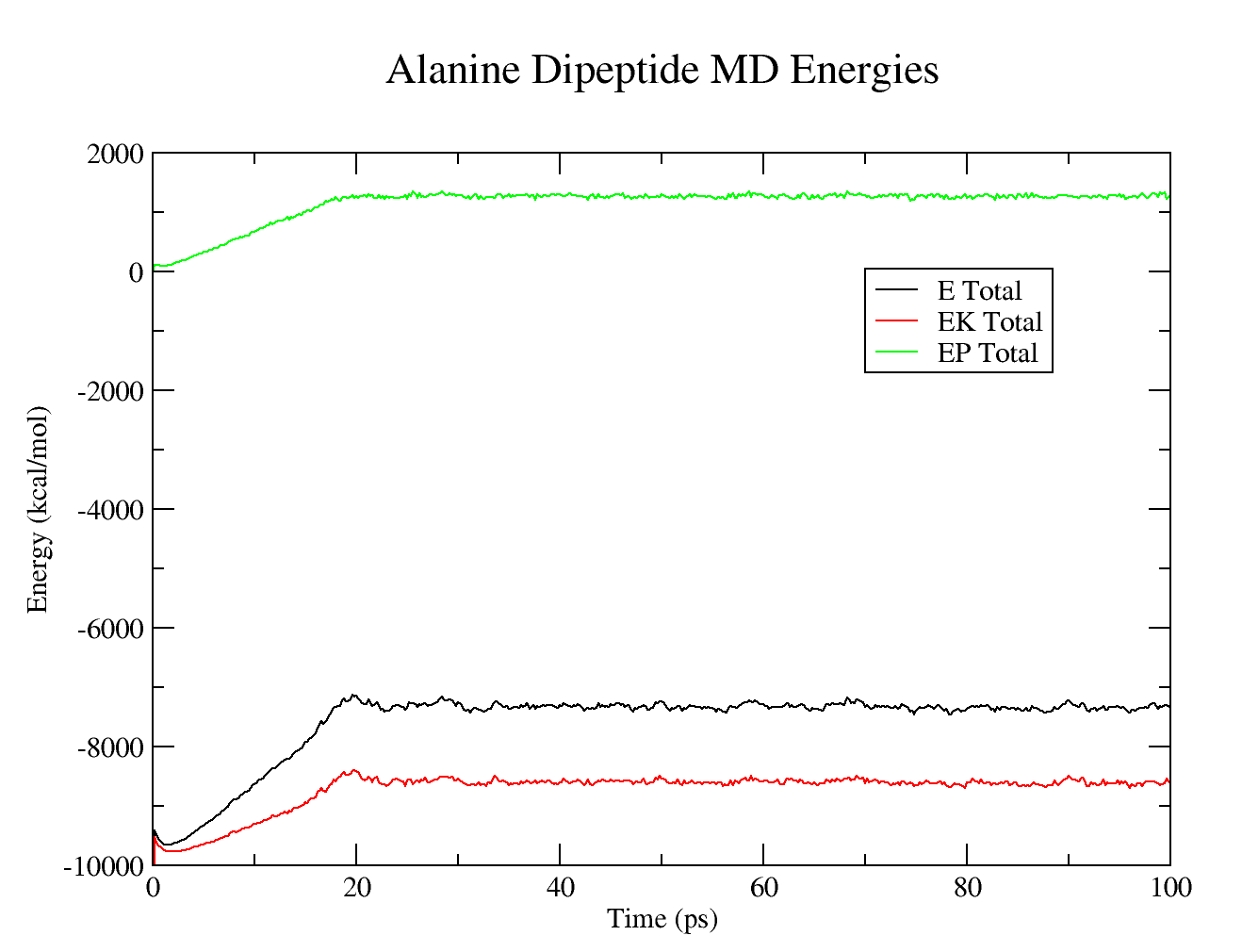
Simple Simulation of Alanine Dipeptide
Recomendado para você
-
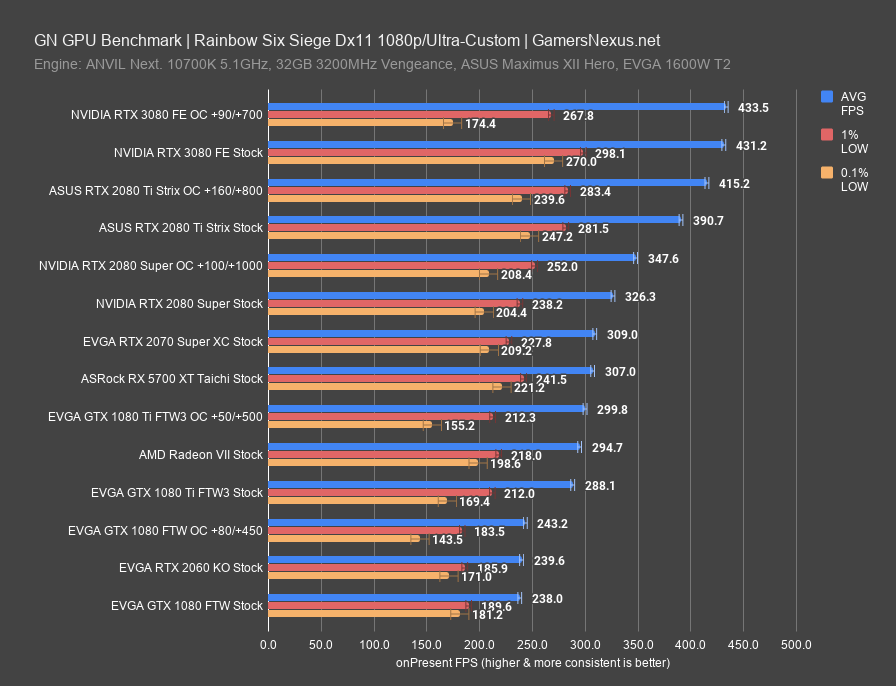 NVIDIA GeForce RTX 3080 Founders Edition Review & GPU Benchmarks21 setembro 2024
NVIDIA GeForce RTX 3080 Founders Edition Review & GPU Benchmarks21 setembro 2024 -
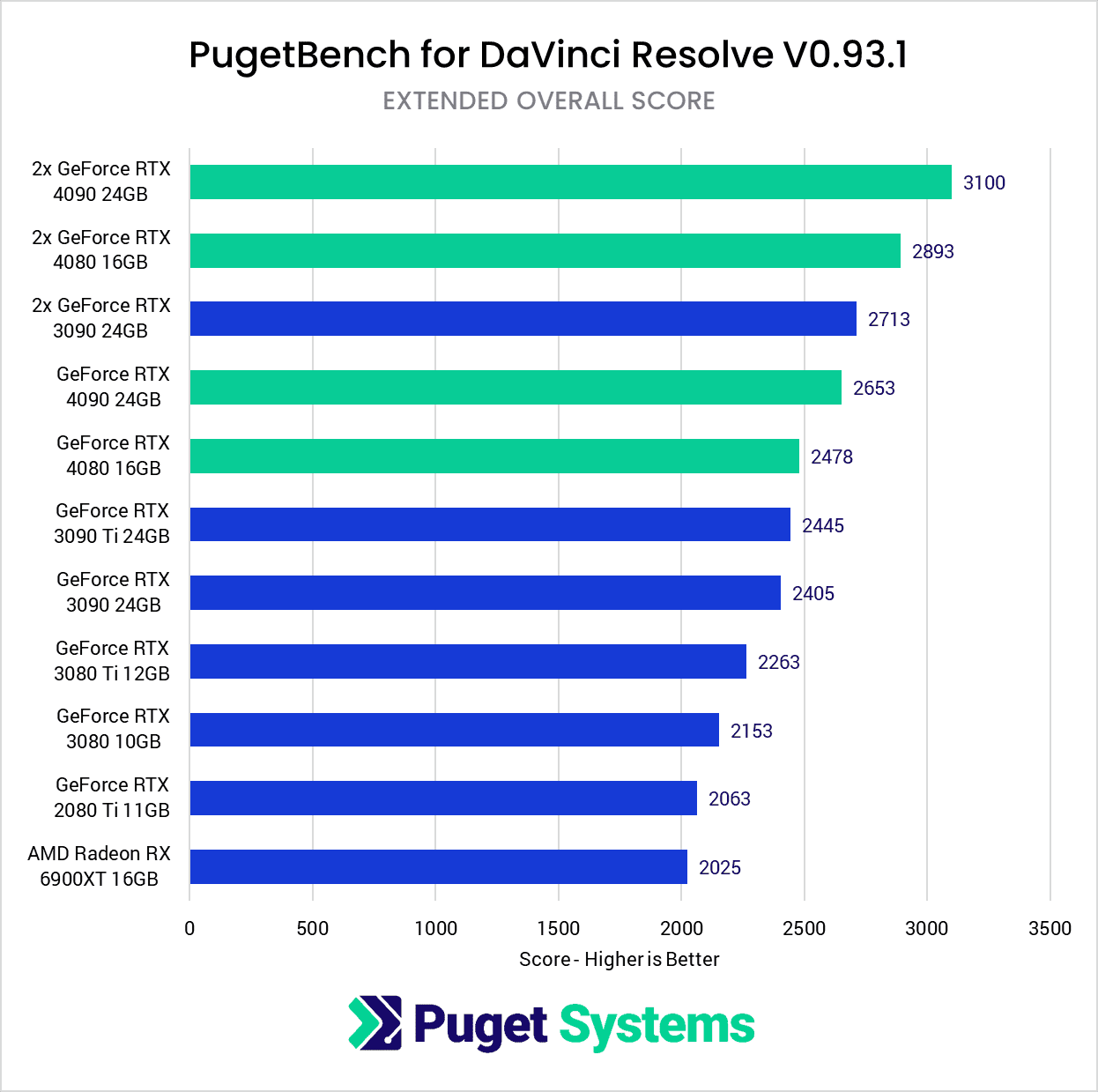 Hardware Recommendations for DaVinci Resolve21 setembro 2024
Hardware Recommendations for DaVinci Resolve21 setembro 2024 -
 PassMark Software - Video Card (GPU) Benchmark Charts21 setembro 2024
PassMark Software - Video Card (GPU) Benchmark Charts21 setembro 2024 -
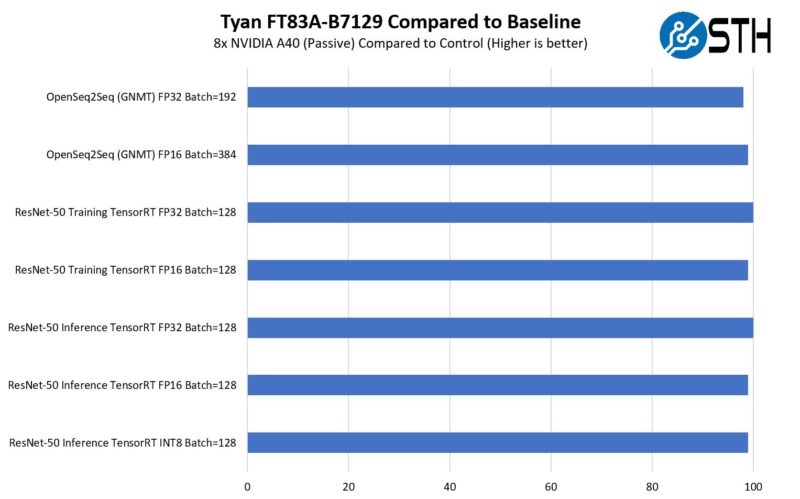 NVIDIA A40 48GB GPU Mini-Review - Page 2 of 2 - ServeTheHome21 setembro 2024
NVIDIA A40 48GB GPU Mini-Review - Page 2 of 2 - ServeTheHome21 setembro 2024 -
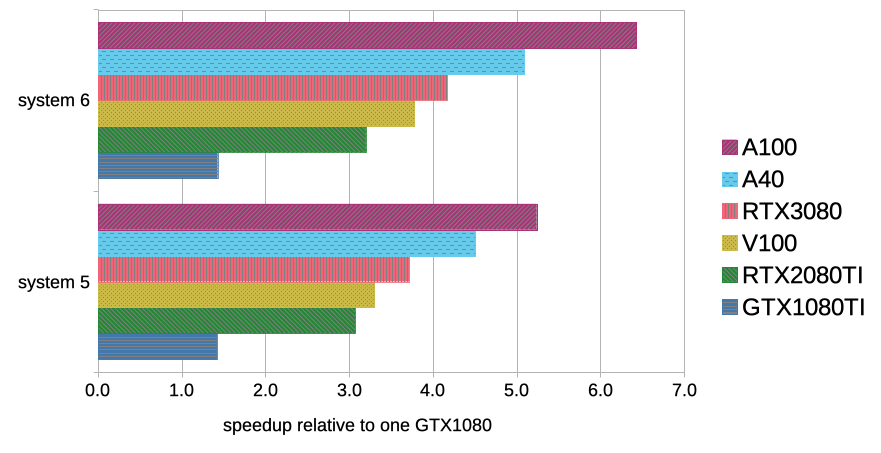 Gromacs performance on different GPU types21 setembro 2024
Gromacs performance on different GPU types21 setembro 2024 -
 Control Benchmark Test & RTX Performance Analysis - Performance21 setembro 2024
Control Benchmark Test & RTX Performance Analysis - Performance21 setembro 2024 -
 Intel Arc A750 performance competes well against the Nvidia RTX 306021 setembro 2024
Intel Arc A750 performance competes well against the Nvidia RTX 306021 setembro 2024 -
 Nvidia GeForce GTX 1050 Ti benchmarks: the fastest budget gaming21 setembro 2024
Nvidia GeForce GTX 1050 Ti benchmarks: the fastest budget gaming21 setembro 2024 -
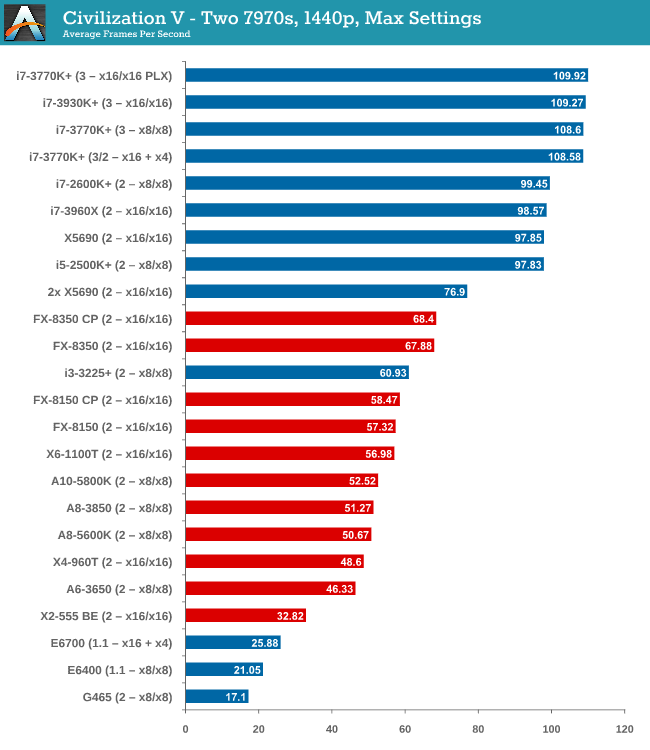 GPU Benchmarks: Civilization V - Choosing a Gaming CPU: Single +21 setembro 2024
GPU Benchmarks: Civilization V - Choosing a Gaming CPU: Single +21 setembro 2024 -
 Intel Benchmarks for Arc A770 Card Suggest It'll Compete With RTX21 setembro 2024
Intel Benchmarks for Arc A770 Card Suggest It'll Compete With RTX21 setembro 2024
você pode gostar
-
 My Beloved Heart Locket GIF - Find & Share on GIPHY21 setembro 2024
My Beloved Heart Locket GIF - Find & Share on GIPHY21 setembro 2024 -
 Conjunto casal Dragon Ball Z Goku e Chi Chi completo - Princesa Urbana - Viva o Encanto21 setembro 2024
Conjunto casal Dragon Ball Z Goku e Chi Chi completo - Princesa Urbana - Viva o Encanto21 setembro 2024 -
 Sudoku Easy 221 setembro 2024
Sudoku Easy 221 setembro 2024 -
 gotoh hitori (bocchi the rock!) drawn by camui110421 setembro 2024
gotoh hitori (bocchi the rock!) drawn by camui110421 setembro 2024 -
TOAA VS ARCHIE SONIC #marvel #narvelcomics #toaa #theoneaboveall21 setembro 2024
-
roblox how to play lone survival controls|TikTok Search21 setembro 2024
-
 blox fruits Personagens de anime, Imagem dragon, Deuses egípcios21 setembro 2024
blox fruits Personagens de anime, Imagem dragon, Deuses egípcios21 setembro 2024 -
 Calça Legging 3D Poliamida Cos Alto – Loja Danna Shop21 setembro 2024
Calça Legging 3D Poliamida Cos Alto – Loja Danna Shop21 setembro 2024 -
 House of the Dragon: O que esperar da série spin-off de Game of Thrones - Revista O Grito! — Cultura pop, cena independente, música, quadrinhos e cinema21 setembro 2024
House of the Dragon: O que esperar da série spin-off de Game of Thrones - Revista O Grito! — Cultura pop, cena independente, música, quadrinhos e cinema21 setembro 2024 -
 10 peças, decoração de bolo de borboleta de papel de simulação de21 setembro 2024
10 peças, decoração de bolo de borboleta de papel de simulação de21 setembro 2024

