DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Por um escritor misterioso
Last updated 20 setembro 2024
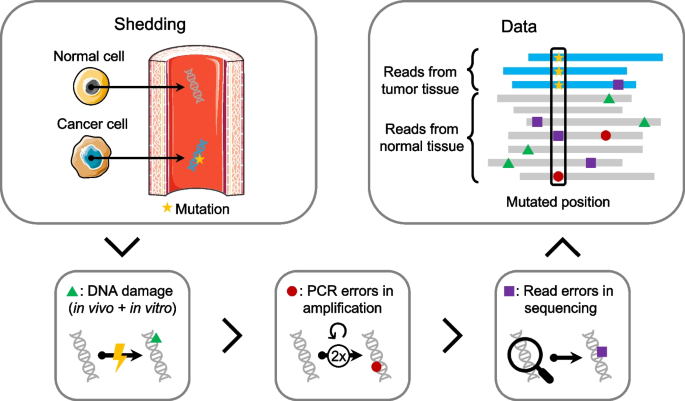
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.

Applications and analysis of targeted genomic sequencing in cancer
PDF) Error Characterization and Statistical Modeling Improves

PDF) DREAMS: deep read-level error model for sequencing data

Discovery of clonal hematopoiesis driver genes a Summary of the
Cancers, Free Full-Text
Genome-wide cell-free DNA mutational integration enables ultra
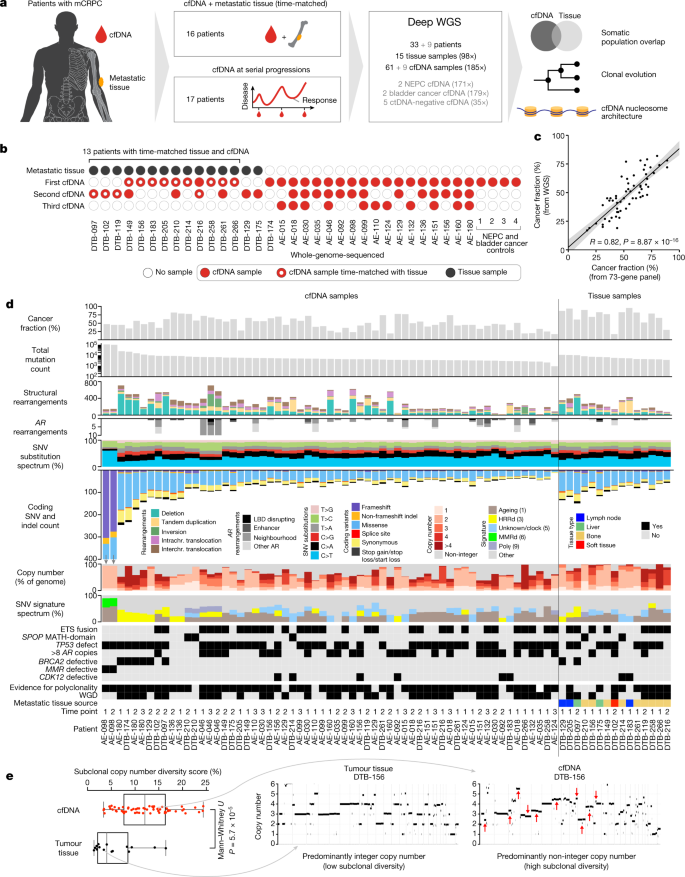
Deep whole-genome ctDNA chronology of treatment-resistant prostate
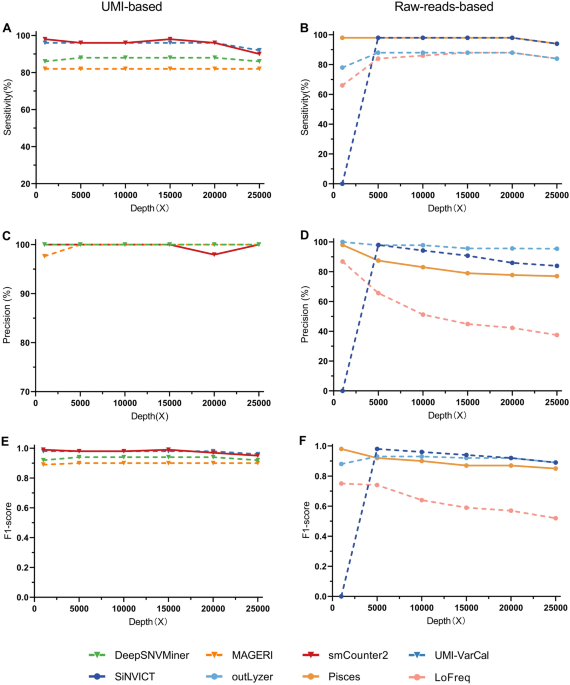
Evaluating the performance of low-frequency variant calling tools
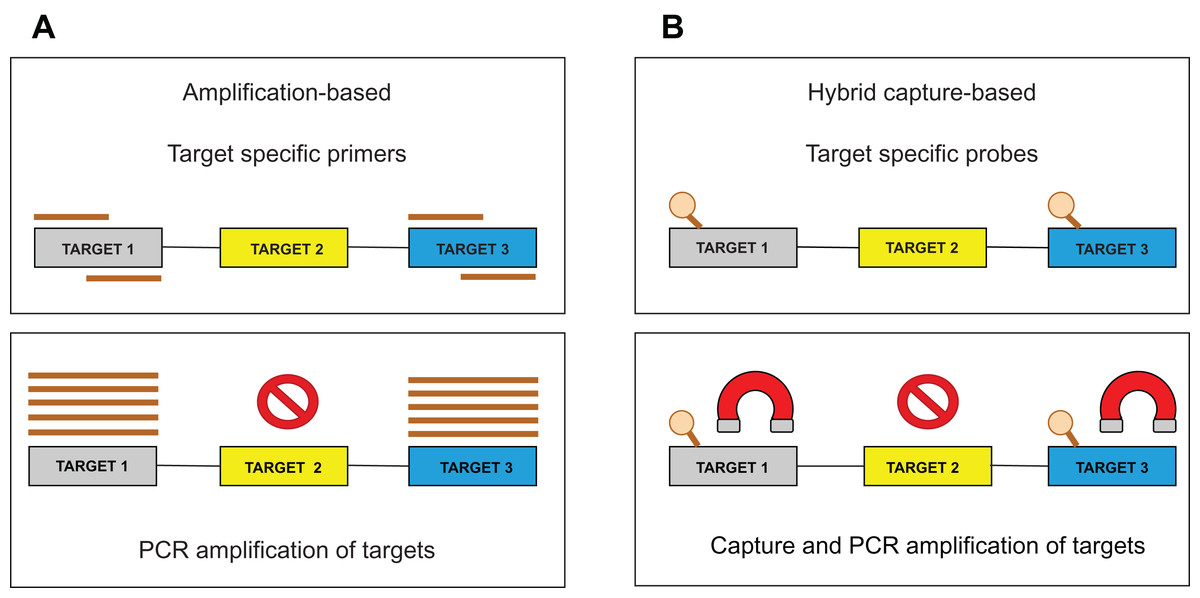
Bioinformatic strategies for the analysis of genomic aberrations

DREAMS: Deep Read-level Error Model for Sequencing data applied to
Recomendado para você
-
![SCP 7143 J THE KNOB [SFW?]](https://i.ytimg.com/vi/qL8GcFVnURE/hq720_2.jpg?sqp=-oaymwEYCI4CEOADSFryq4qpAwoIARUAAIhC0AEB&rs=AOn4CLCHvLM5-ZlLxt7xHLsYYfAnNKRhFA) SCP 7143 J THE KNOB [SFW?]20 setembro 2024
SCP 7143 J THE KNOB [SFW?]20 setembro 2024 -
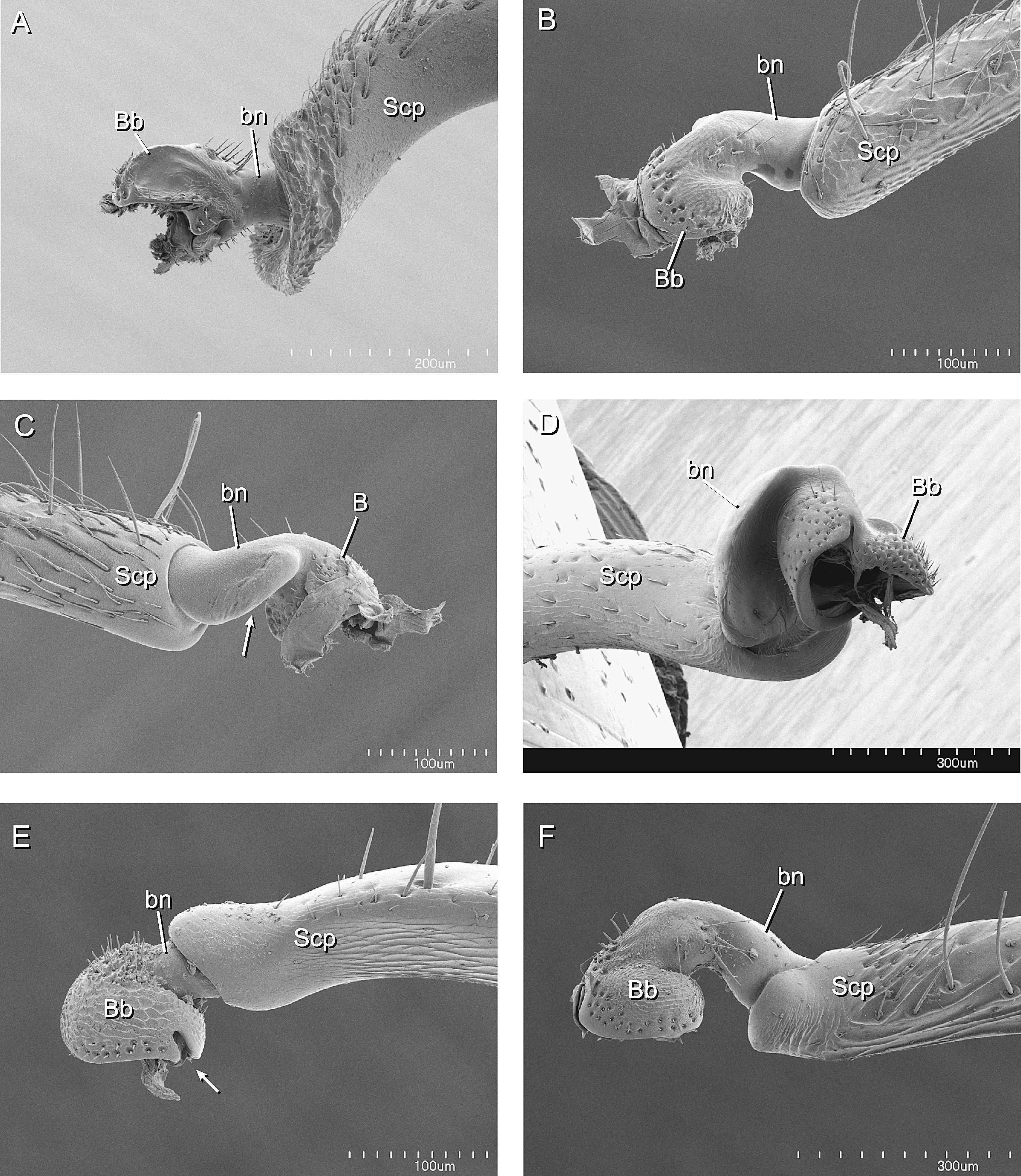 A Phylogenetic Analysis of Ant Morphology (Hymenoptera: Formicidae) with Special Reference to the Poneromorph Subfamilies20 setembro 2024
A Phylogenetic Analysis of Ant Morphology (Hymenoptera: Formicidae) with Special Reference to the Poneromorph Subfamilies20 setembro 2024 -
 S I M P SCP Foundation (RP) Amino20 setembro 2024
S I M P SCP Foundation (RP) Amino20 setembro 2024 -
![REDACTED] — I would fuck the shit out of that doorknob.](https://64.media.tumblr.com/cf4c7805a721dd20b047ecbd0ae466d5/tumblr_pnvhf1VWpn1y0mr9no1_500.jpg) REDACTED] — I would fuck the shit out of that doorknob.20 setembro 2024
REDACTED] — I would fuck the shit out of that doorknob.20 setembro 2024 -
 Emolog by Kyungmoo Min20 setembro 2024
Emolog by Kyungmoo Min20 setembro 2024 -
 SCP-5417-J: The Deadly Nackle20 setembro 2024
SCP-5417-J: The Deadly Nackle20 setembro 2024 -
open_science_fellowship_project/resources/example_data/derivatives/freesurfer/sub-01/scripts/recon-all.local-copy at master · PeerHerholz/open_science_fellowship_project · GitHub20 setembro 2024
-
 Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum20 setembro 2024
Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum20 setembro 2024 -
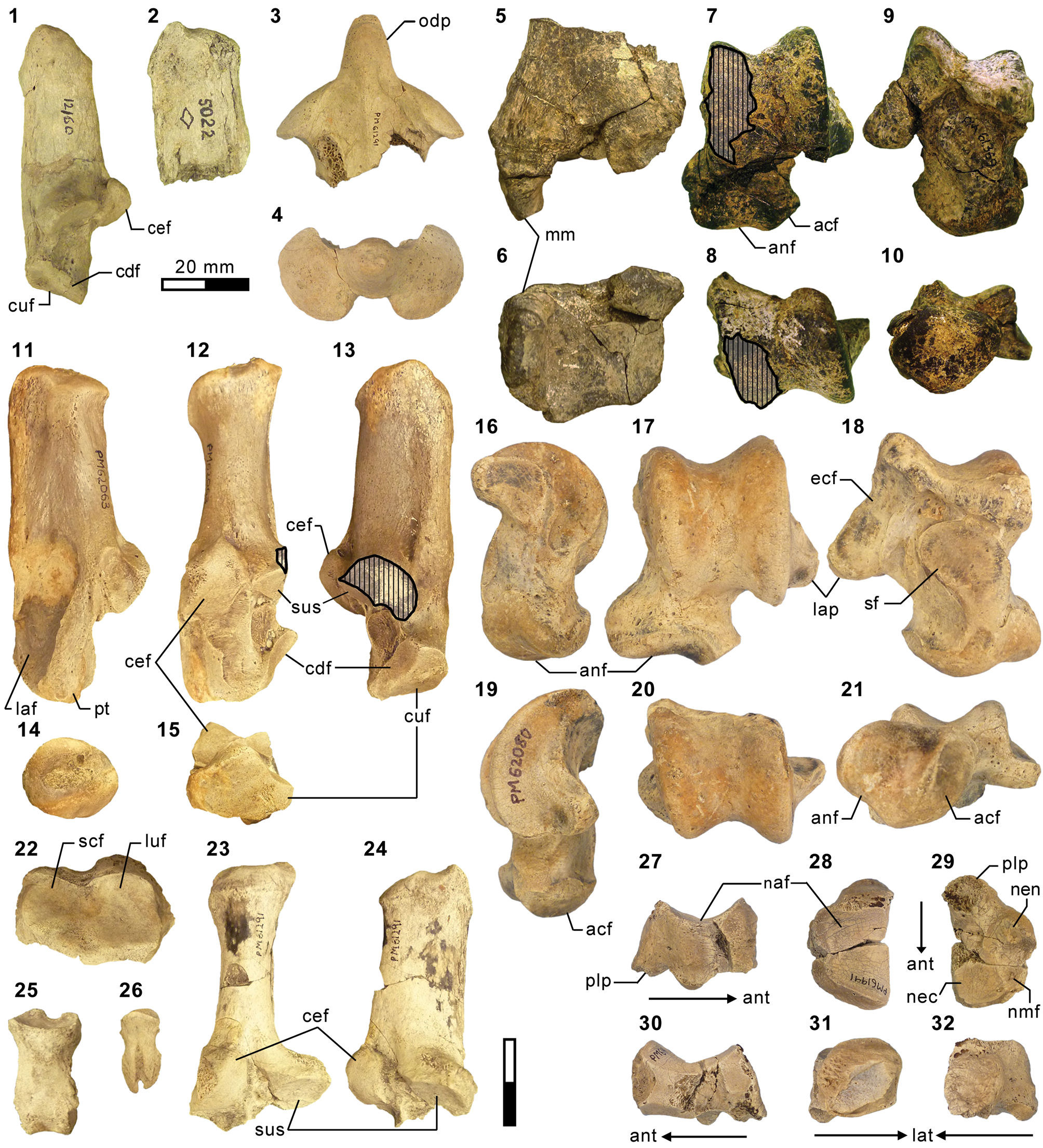 Carnivorous mammals from the middle Eocene Washakie Formation, Wyoming, USA, and their diversity trajectory in a post-warming world, Journal of Paleontology20 setembro 2024
Carnivorous mammals from the middle Eocene Washakie Formation, Wyoming, USA, and their diversity trajectory in a post-warming world, Journal of Paleontology20 setembro 2024 -
Transportador de Recorte, PDF, Gear20 setembro 2024
você pode gostar
-
 Os 15 Melhores Jogos De Multiplayer Online Para Android 202020 setembro 2024
Os 15 Melhores Jogos De Multiplayer Online Para Android 202020 setembro 2024 -
 LEGO: Harry Potter Com Hedwig - Harry Potter - Toyshow Tudo de20 setembro 2024
LEGO: Harry Potter Com Hedwig - Harry Potter - Toyshow Tudo de20 setembro 2024 -
 How to use Xbox Cloud Gaming on your Android device - Phandroid20 setembro 2024
How to use Xbox Cloud Gaming on your Android device - Phandroid20 setembro 2024 -
 Ryan Reynolds Shared A Meme About The Last Two Years & It Of Course Featured A Car Crash - Narcity20 setembro 2024
Ryan Reynolds Shared A Meme About The Last Two Years & It Of Course Featured A Car Crash - Narcity20 setembro 2024 -
 Light Novel Edition Kurumi Tokisaki: Wedding Dress Ver.20 setembro 2024
Light Novel Edition Kurumi Tokisaki: Wedding Dress Ver.20 setembro 2024 -
 Segunda temporada da série One Piece é confirmada pela Netflix - Folha BV20 setembro 2024
Segunda temporada da série One Piece é confirmada pela Netflix - Folha BV20 setembro 2024 -
 Lena 86 Eighty-Six SH Figuarts Figure20 setembro 2024
Lena 86 Eighty-Six SH Figuarts Figure20 setembro 2024 -
 Blister(pacote)Quadruplo Pokemon Origem Perdida 25 Cartas (aleatório) em Promoção na Americanas20 setembro 2024
Blister(pacote)Quadruplo Pokemon Origem Perdida 25 Cartas (aleatório) em Promoção na Americanas20 setembro 2024 -
 Prefeitura Municipal de Pompéu - Campeonato Infantil de Futsal e o Campeonato de Bairros20 setembro 2024
Prefeitura Municipal de Pompéu - Campeonato Infantil de Futsal e o Campeonato de Bairros20 setembro 2024 -
 3D printer Cyan BLUE ROBLOX: Rainbow Friends 3d model • made with Ender 3 pro・Cults20 setembro 2024
3D printer Cyan BLUE ROBLOX: Rainbow Friends 3d model • made with Ender 3 pro・Cults20 setembro 2024
